What Will You Research?
2025 Big Data Summer Institute Projects
BDSI participants are split into research teams of 10 students to focus on a specific biomedical or public health application. Each team has at least one faculty mentor and one graduate student research assistant to facilitate the project work. Participants are assigned to research groups based on indicated preferences and skill sets. The specific aims and datasets are chosen closer to the start of the program.
 Project Group One: Cancer Data Science
Project Group One: Cancer Data Science
The Cancer Data Science group will delve into statistical, computational, and mathematical questions that arise in cancer research. The research project will involve an application to advancing cancer prevention and care. Examples include developing predictive models to assess recurrence risk based on clinical, genomic, or pathological data; investigating spatial and temporal dynamics of tumor evolution using genomic and medical imaging data; leveraging clinical data to identify biomarkers associated with tumor initiation and progression, and much more. Student teams will initiate novel research questions using the provided data sources and conduct in-depth analysis to explore these questions. This immersive experience will teach students valuable skills in data manipulation, statistical computing, and data visualization. Within this research group, students will have a chance to engage with members of the UM Cancer Data Science group and learn to apply advanced statistical methods, such as survival analysis, machine learning, and spatial data analysis.
Project Mentors: Dr. Nicholas Hartman
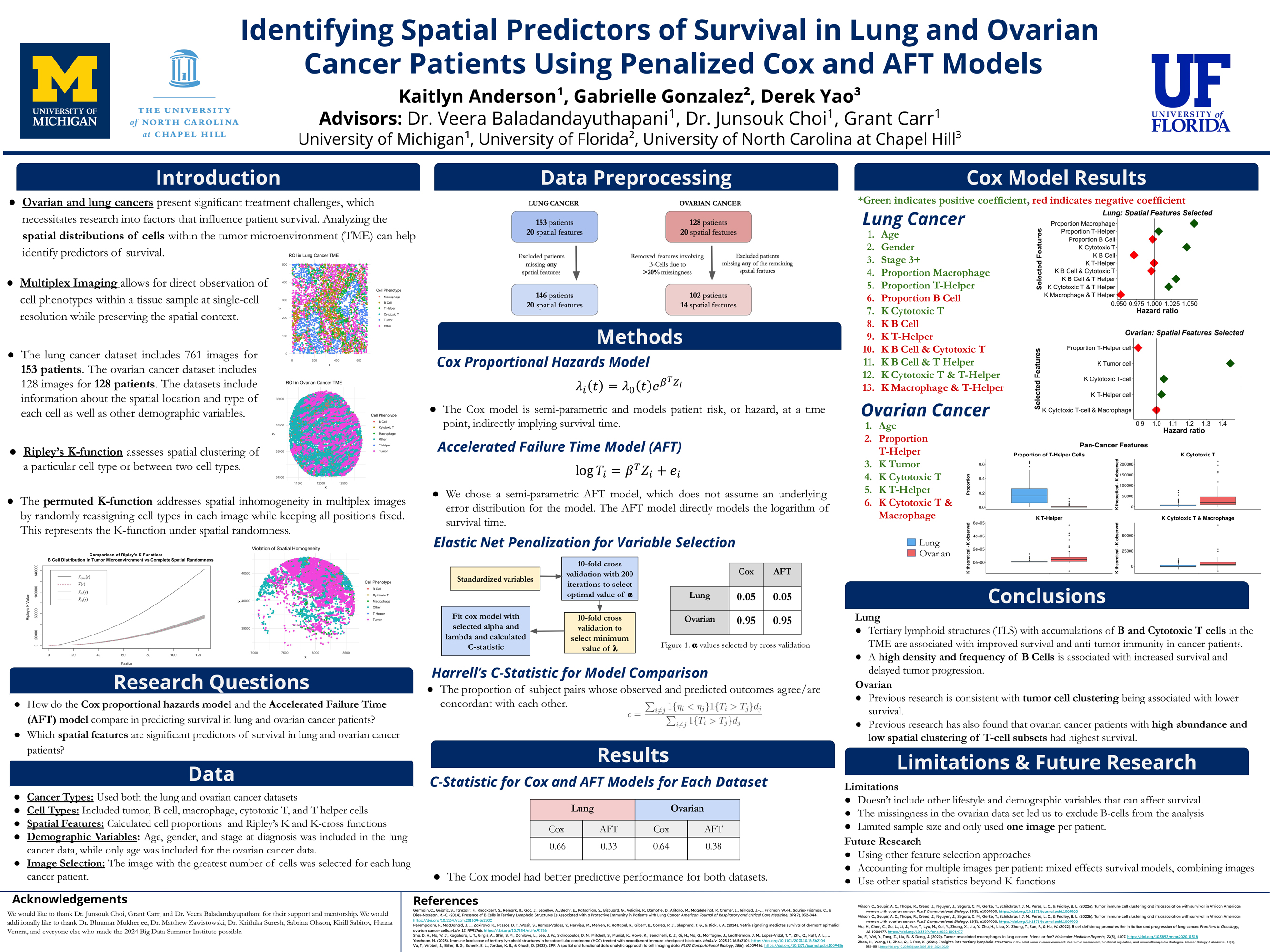
Examples of Past Projects
 Project Group Two: Machine Learning in Healthcare Data
Project Group Two: Machine Learning in Healthcare Data
Statistics for Trustworthy Machine Learning: An accepted pathway to affirm a discovery is by replicating the experiment on data. Despite use of machine learning (ML) across scientific fields, an alarmingly high proportion of ML findings do not hold up in new datasets: this has led to the pressing replication crisis. To gauge chances of replication, researchers need tools to understand uncertainties in ML findings.
Multiple projects under the "Statistics for Trustworthy ML" theme will apply ML to model clinical data from imaging and genomics. These projects will investigate the potential of related findings from a replication perspective, and apply some recent tools in selective inference for uncertainty estimation. Students will gain experience in modeling large and complex datasets, and will be introduced to selective inference for improving replication.
Project Mentors: Dr. Katherine Brumberg, Dr. Rahul Ladhania
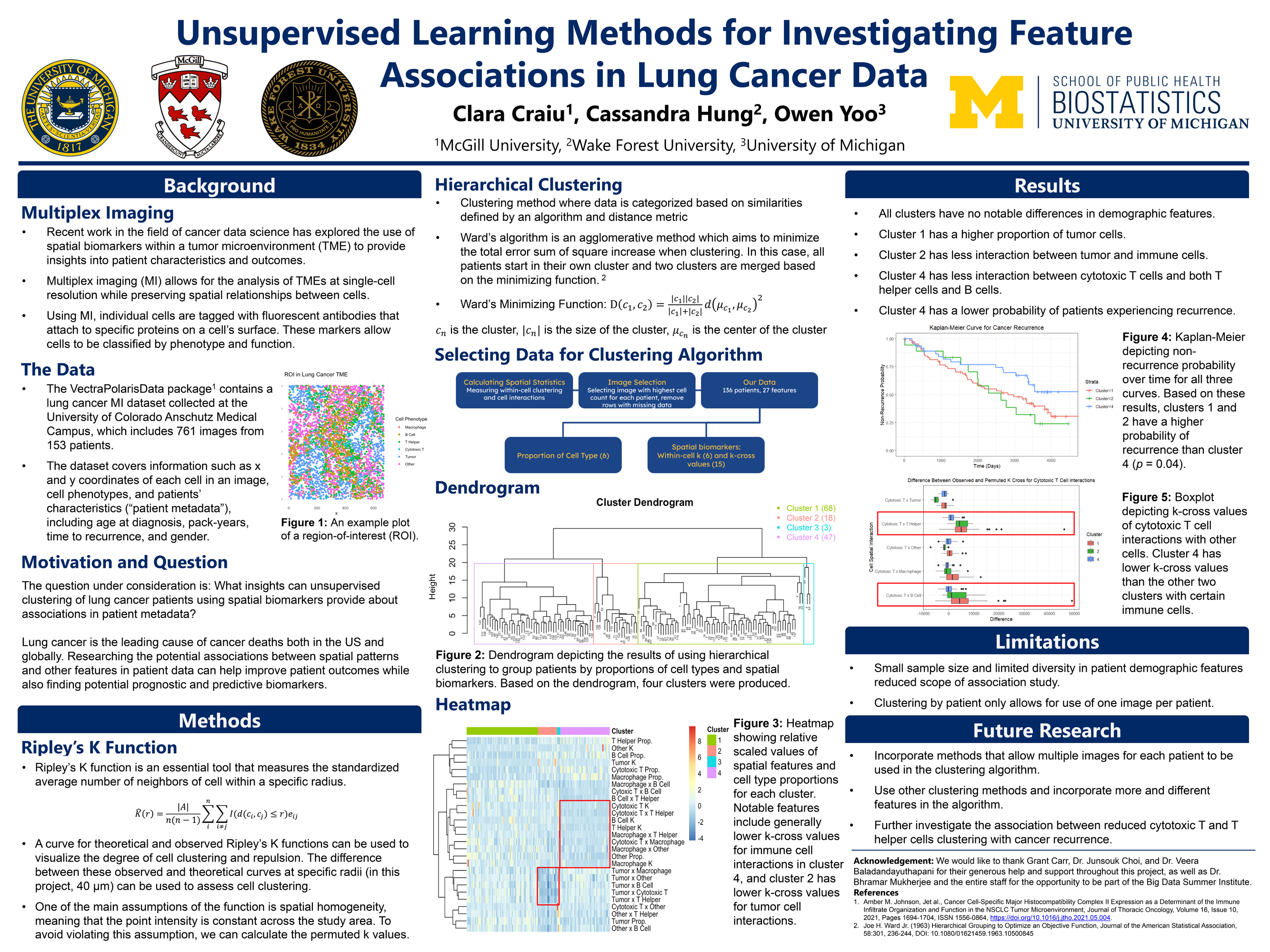
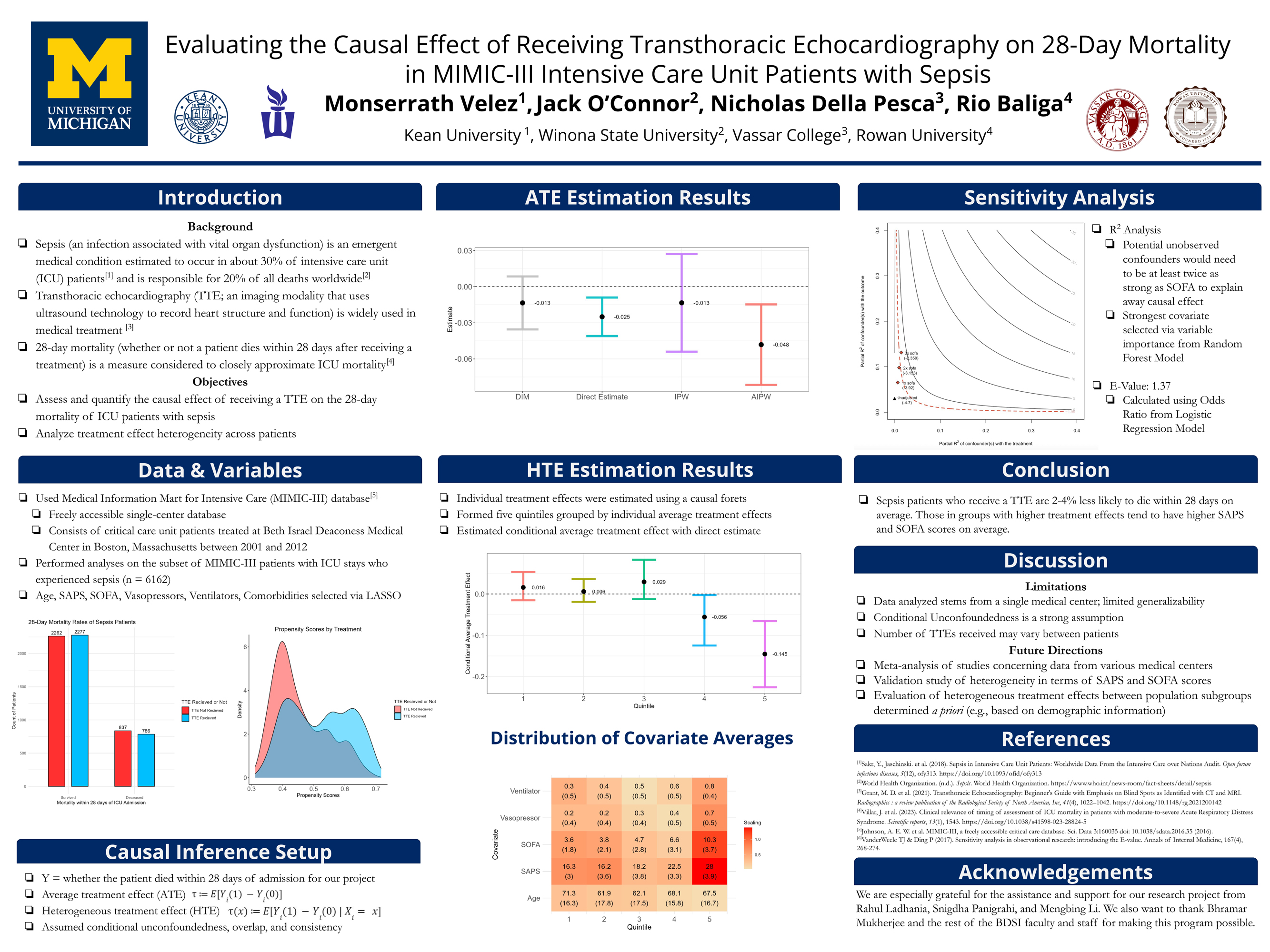
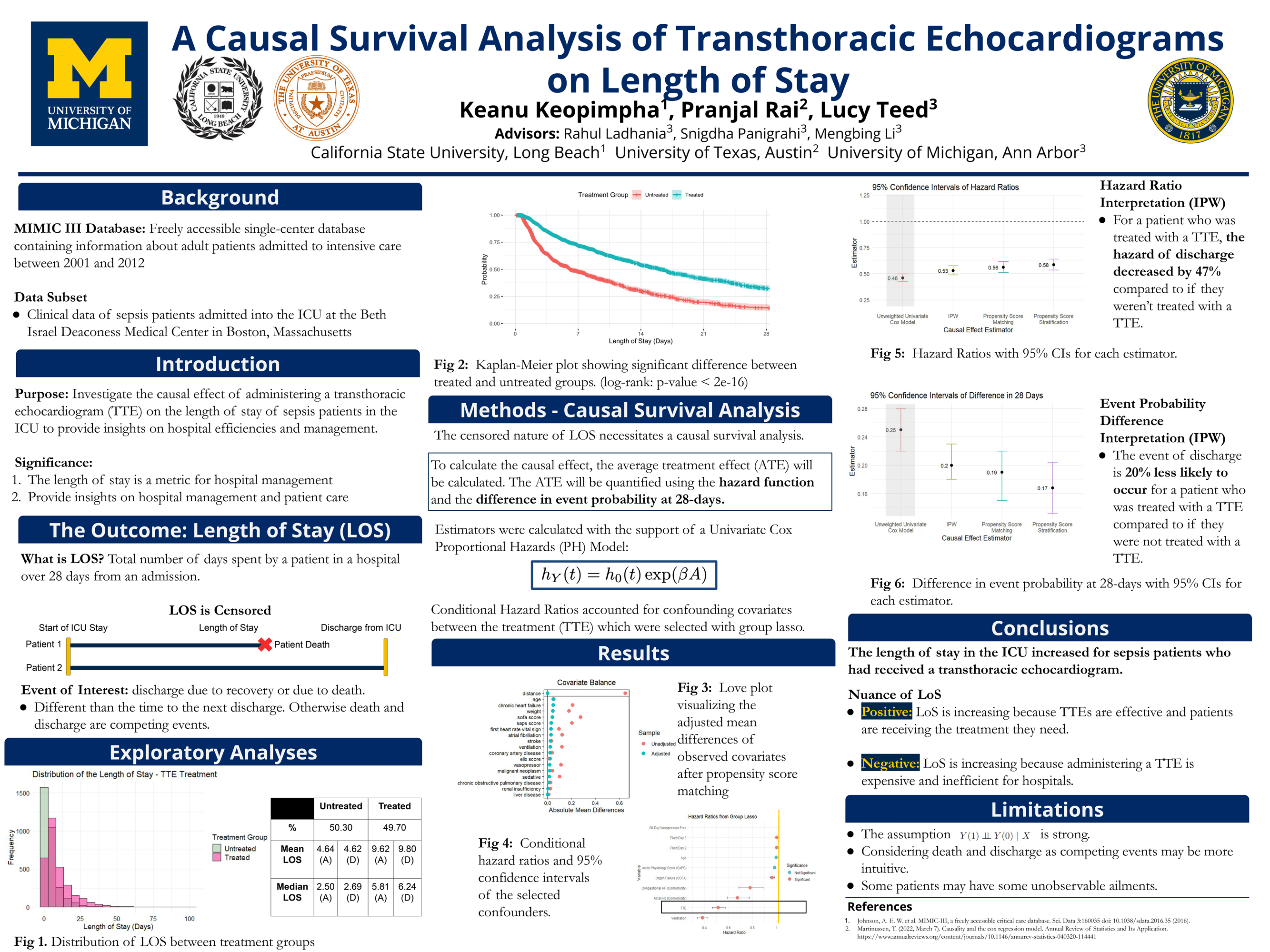
Examples of Past Projects
 Project Group Three: Genomics
Project Group Three: Genomics
The Genomics group will have multiple available projects connecting a health-related question to a large-scale genomic dataset, for example whole-genome Single Nucleotide Polymorphism data, single-cell RNA sequencing data or epigenetic methylation data. Students will form teams for a deep dive analysis on their specific project of interest with opportunity for open-ended exploration. Students will gain hands-on computing experience and valuable data manipulation skills working with the large genomic data files. We will apply many classical statistical techniques, learn about integration of complementary genomic data sources and explore machine learning and specialized genomic analysis methods.
Project Mentors: Dr. Matt Zawistowski, Dr. Xiang Zhou